Natural Language Processing (NLP)
1. Natural Language Processing
The roots of modern NLP systems can be traced back to the concept of “Language as a Science” developed in the early 1900s that started with linguistics and later expanded to computers and other fields. NLP came to the fore after World War II based on the need for a machine that could automatically translate from one language to another. Then in 1950, interest in NLP began in earnest with the publishing of Alan Turing’s seminal paper “Computing Machinery and Intelligence.” The paper proposed that a computer that could converse with human beings without them realizing they were talking to a machine could be considered intelligent.
Today, NLP is everywhere. Though digital voice assistants are the most ubiquitous real-world application of NLP, the concept itself encompasses both speech and text and is used in a variety of applications including search, email spam filtering, online translation, grammar- and spell-checking, and more.
Large Language Models in Biomedical NLP
Natural Language Processing (NLP) addresses a longstanding knowledge gap in life sciences research and drug discovery, namely that of integrating vast volumes of unstructured textual data embedded in scientific articles, clinical trial reports, electronic health records, etc.
In recent years, the emergence of Large Language Models (LLMs), such as ChatGPT, has shown the potential to revolutionize life sciences research with powerful new natural language processing capabilities that could accelerate scientific discovery and enhance the efficiency of drug development. These models have demonstrated that with domain-specific pretraining they can outperform conventional small molecule drug discovery processes as well as help improve or design new antibodies.
However, there are still a number of challenges that have to be addressed — including the real-world consequences of generative AI’s tendency to hallucinate and the interpretability/explainability issues associated with black box LLMs — before these models can be scalably applied in the life sciences industry.
The most significant challenge though is conceptual.
The learning capabilities of LLMs are based on identifying co-occurrence patterns from a huge corpus of text. Though this approach to language understanding based on the probability of word co-occurrence may well capture syntax and semantics, there is a heated debate on whether massive statistical models can enable true human-like understanding. Also, since LLMs are probabilistic in nature, they can generate a range of possible outputs, each with its own probability of being correct, for any given input. In the case of biomedical research, LLM-generated results will require additional verification and validation for accuracy.
As a distinct subset of NLP, LLMs have demonstrated their potential in a range of biomedical language processing tasks. However, they are still predominantly focused on literal language use and the ability of statistical language models to account for pragmatic inferences, contextually-modulated meaning, and complex analogies continues to be the focus of research interest.
The best way forward is with hybrid NLP, a design philosophy that combines state-of-the-art NLP technologies, components, and models into one integrated outcome-focused pipeline that enhances productivity, accuracy, and interpretability.
Read more on our blog: From words to meaning: Exploring semantic analysis in NLP
1.1. What is Natural Language Processing?
Natural-language processing (NLP) refers to the automated computational processing technologies that convert natural language text or audio speech into encoded, structured information, based on an appropriate ontology. In the context of biomedical literature, the structured information can then be used to classify a body of textual information, as in “related to laparoscopic cholecystectomy,” or to extract more refined insights such as participants, procedures, findings, etc.
Read more on our blog: AI, ML, DL and NLP: An overview
1.2. How does NLP work?
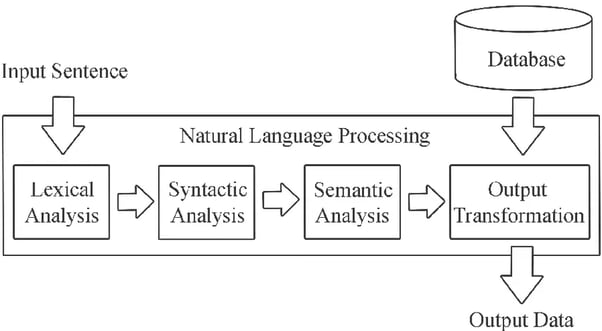
SOURCE: AI Multiple
In simple terms, NLP works through machine learning (ML) systems that store words and information on the ways they relate to each other. ML engines use grammatical rules and real-world linguistic patterns to process words, phrases and sentences and extract meaning, context and intent. However, NLP derives from a broad range of techniques for interpreting human language, including rules-based and algorithmic approaches, statistical and machine learning methods, and deep learning and neural networks.
2. NLP tasks
All NLP Tasks are considered to be generation tasks with the three main categories being classification, unconditional generation, and conditional generation.
Here’s a brief overview of some of the tasks from within each of those categories.
2.1. Speech Recognition
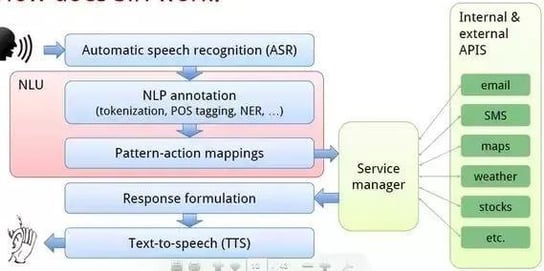
Source: Researchgate
Speech recognition, also known as automatic speech recognition (ASR), computer speech recognition, or speech-to-text, is the ability of a program to process human speech and convert it into readable text.
ASR systems can be classified based on utterances (connected words, continuous speech etc.), speakers (speaker independent, speaker adaptive etc.), or even vocabulary (small to very large vocabulary).
Speech recognition systems typically follow four steps: analyze audio, deconstruct, digitize, and use an algorithm to create the most suitable text representation.
2.2. Part of Speech Tagging
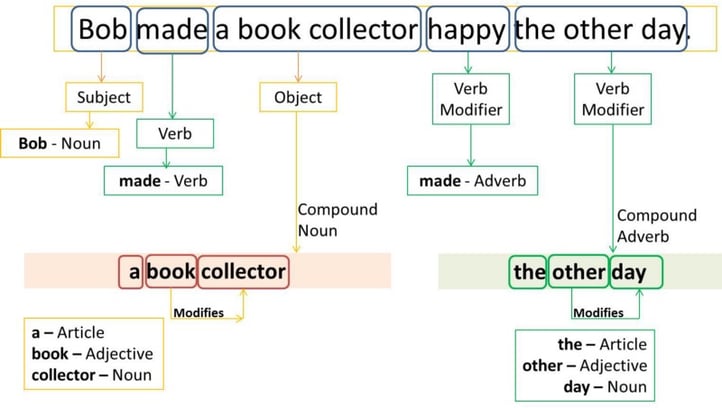
Source: freeCodeCamp
Part of Speech (POS) describes the grammatical function of a word. There are typically 8 parts of speech — noun, verb, pronoun, preposition, adverb, conjunction, participle, article — that are relevant to NLP. POS tagging, also known as grammatical tagging, is the process of automatically assigning POS tags to words in a sentence.
Most POS tagging approaches fall under one of three categories; rule-based, stochastic, or transformation-based tagging.
Rule-based POS tagging relies on a dictionary or lexicon to generate tags with additional hand-written rules used to identify the correct tag from a set of possible tags for a word.
Stochastic POS Tagging refers to any model that includes frequency or probability statistics to determine the most appropriate tags based on the probability of occurrence or the frequency of their association with a word in a training corpus.
Transformation-based tagging is based on transformation-based learning and incorporates features from both previous approaches. Just as in rule-based tagging, it relies on rules that associate tags to words – and like in stochastic tagging, it applies ML techniques to automatically deduce rules from data.
2.3. Word Sense Disambiguation
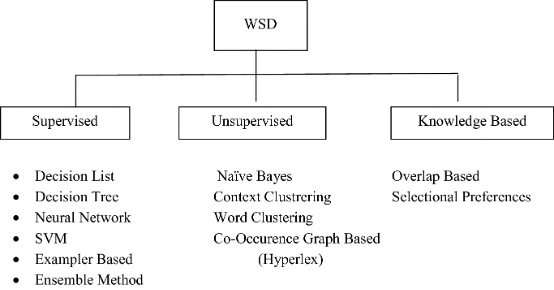
Source: Springer
Word sense disambiguation (WSD) is the process of finding the meaning of a word that is most suitable to the context. WSD seeks to resolve one of the most pervasive linguistic challenges in NLP – polysemous words or words that have multiple related meanings. The primary purpose of WSD, therefore, is to clarify the contextually appropriate meaning for polysemous words.
There are four main ways to implement WSD; Dictionary- and knowledge-based, supervised, semi-supervised, and unsupervised methods.
2.4. Named Entity Recognition
Named Entity Recognition (NER) is an NLP technique for identifying and extracting essential entities from text-based documents. Typical essential entities include names of people, locations, organisations, monetary values, etc. However, named entities can cover a wide number of categories, such as unit, type, quantity, occupation, ethnicity etc., and depend on the NLP requirement.
Biomedical named entity recognition (BioNER) is a specialised field dealing with the extraction of biomedical entities from scientific texts. The automated and accurate identification of entities from a rapidly growing library of literature can streamline and accelerate several downstream tasks in biomedical research.
Deep learning methods and end-to-end neural networks have been used quite successfully to automatically extract relevant features from biomedical text. The deep learning methods typically used for NER are classified into four categories: single neural network-based, multitask learning-based, transfer learning-based, and hybrid model-based methods. These methods can be used for BioNER applications across multiple domains.
2.5. Coreference Resolution
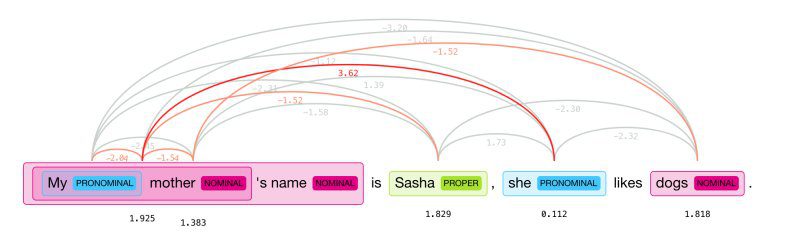
Source: Hugging Face
Coreference resolution (CR) is the task of finding all linguistic expressions, or mentions, in a given text that refer to the same real-world entity. Once all the mentions have been identified and grouped, they can be replaced with words that provide context. For instance, a sentence could introduce a person by name and then subsequently use pronouns to refer to the same person. CR determines if two mentions refer to the same discourse entity in the discourse model.
2.6. Sentiment Analysis
Sentiment analysis is an analytical technique that determines the emotional meaning of communications. Sentiment Analysis identifies whether a message is positive, negative or neutral and interprets what people are feeling via their language.
Some of the most popular methods of sentiment analysis include standard, fine-grained and aspect-based sentiment analysis. The standard approach provides a broad interpretation of the overall tone of communication. The fine-grained model accounts for a more elaborate range of polarity. And the aspect-based approach delivers a more precise interpretation of sentiment based on particular attributes and components.
2.7. Natural Language Generation
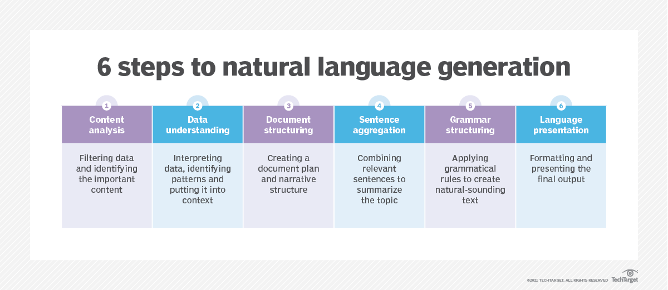
Source: TechTarget
Natural language generation (NLG) is the process of transforming data into natural language. The process involves applying statistical techniques to analyse large datasets of structured information to generate natural-sounding sentences. NLG systems can automatically turn numbers in a spreadsheet into data-driven narratives or even generate entire articles or responses.
NLG is a six-stage process that includes content analysis, data understanding, document structuring, sentence aggregation, grammatical structuring, and language presentation.
2.8. Relation Extraction
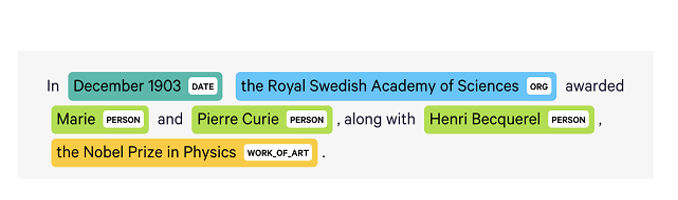
Source: Open Data Science
Relation Extraction (RE) is the task of predicting attributes and relations for entities in a sentence. It is a key component for building relation knowledge graphs and is used in several NLP applications including structured search, sentiment analysis, question answering, and summarization. End-to-end relation extraction can help identify named entities and extract relations between them.
For instance, text mining is increasingly being used in the biomedical domain to automatically organise information from large volumes of scientific literature. In this context, relation extraction aims to identify designated relations among biological entities in literature. RE can also facilitate the extraction of semantic relations between different biomedical entities such as protein and protein, gene and protein, drug and drug, and drug and disease.
Read more on our blog: Knowledge graphs & the power of contextual data
3. NLP in real life
3.1. NLP Use Cases in Life Sciences
In the biomedical and life sciences domain, NLP opens up access to data sources, like scientific journals and medical/clinical data, that were previously incompatible with conventional data analytics frameworks. As a result, it has catalysed a gamut of real-world applications in novel drug target intelligence, biomarker discovery, safety case processing, clinical trial analytics, medical affairs insights and clinical documentation improvement.
Here’s a brief overview of how NLP is being leveraged in the life sciences.
Clinical natural language processing
Though clinical records have predominantly moved on from paper to codify valuable information in EHRs, much of the information related to real-world clinical practices still appear as unstructured narrative free-text. This has given rise to a specialised research field called clinical natural language processing (cNLP) to explore clinically relevant information contained in EHRs. cNLP systems have transformed the scope and scale of the utilisation of unstructured free-text information in EHRs, thereby providing valuable insights into clinical populations, epidemiology trends, patient management, pharmacovigilance, and optimisation of hospital resources.
Competitive intelligence from patent literature
Pharma R&D can benefit significantly by extracting competitive intelligence from publicly available patent literature. One pharma major leveraged NLP text mining to automatically extract information related to four main entities, from three major patent registries, and update this data every week.
Disease diagnosis with NLP
Most health systems track patients using codes that are primarily created for billing and are therefore not particularly useful for clinical care or research. This creates a huge challenge in identifying patients with complex conditions and in studying the disease, tracking practice patterns, and managing population health. However, researchers at a large US healthcare provider were able to train an NLP model to automatically sort through over a million EMRs to identify abbreviations, words and phrases associated with aortic stenosis. In a matter of minutes, the NLP algorithms were able to identify nearly 54,000 patients with specific conditions.
NLP for patient stratification
A leading biopharma company was able to focus the patient stratification process by using NLP on EMR and imaging data. By capturing data on 40 different elements related to a number of variables, including demographics, clinical outcomes, clinical phenotypes, etc., researchers were able to identify four patient groups with substantial differences in one- and two-year mortality and one-year hospitalisations. By using insights from the NLP-based analysis the company was able to improve clinical trial design, identify unmet needs, and develop better therapeutics.
NLP advances precision medicine research
Precision medicine transfers the focus of treatment from the average patient to the individual patient. However, developing a personalised medical approach requires substantial volumes of disparate data that need to be analysed in a multi-scale context. A top medical school in the US has adopted NLP tools to pull key information regarding diagnoses, treatments, and outcomes from EHRs.
Read more on our blog: Improving drug safety with adverse event detection using NLP
3.2. BioNLP Case Studies
NLP-ML system for analyzing clinical notes
During the pandemic, the healthcare authority in the Canadian province of Alberta launched a free telehealth service that allowed patients and caregivers to speak directly to rehabilitation clinicians and professionals about the impact of the pandemic on chronic musculoskeletal, neurological, and other conditions. The service was designed to provide callers with assistance regarding services available in their location, condition-specific exercises, self-management advice etc. For every call, clinical notes containing detailed patient information were entered into an online charting platform. Apart from patient demographics, these call notes consisted of several layers of unstructured data that included patient history of diagnoses, medications, and existing symptoms, details about the ensuing discussion including causes, over-the-phone assessment and action taken by the advisor and finally, details about advice/service referrals provided to the patient.
An NLP-ML system was designed for the automated pre-processing of these clinical notes and for modelling and analyzing the collected data. Preliminary results have shown that the NLP system was capable of accurately identifying salient keywords within the clinical notes.
NLP for rapid response to emergent diseases
Conventional bioinformatics largely relies on structured data and preexisting knowledge models. But this approach does not work in the context of novel diseases with no preexisting knowledge models. COVID-19 presented an opportunity to test the hypothesis that NLP technologies can enable the conversion of unstructured text to novel knowledge models. Researchers designed a study to evaluate the value that information from clinical text could add to the response to an emergent disease.
The focus was on COVID-19 infections in high blood pressure patients and the effect of long-term treatment with calcium channel blockers on outcomes. The study used two sources of information: one where data was solely from structured EHRs and the other on data from structured EHRs and text mining.
According to the results of the study, text mining was able to augment statistical power sufficient enough to change a negative result to a positive one. When compared to the baseline study with structured data, the NLP study saw a steep increase in the number of patients available for inclusion, the amount of available information on medications and the amount of additional phenotypic information. The conclusion was that supplementing conventional structured data approaches with information from the NLP pipeline would increase the sample size sufficiently enough to see treatment effects that were not previously statistically detectable.
NLP to detect virus mutations
Viral escape is the ability of viruses to mutate thereby not only evading neutralizing antibodies but also impeding vaccine development. Researchers at MIT have now developed a novel approach to model viral escape based on models originally developed to analyze language.
The basic light bulb idea is that the immune system interprets a virus the same way humans interpret a sentence. The team used the linguistic concepts of grammar and semantics to interpret the characteristics and mutations of a virus. In linguistic terms, the grammatical correctness of a virus determines its evolutionary ability to infect a host. Similarly, mutations are analogous to semantics in that a virus that has altered its surface proteins to become invisible to antibodies is said to have altered its meaning. So in essence a successful virus that can change semantically without compromising grammatical correctness.
The research team trained an NLP model on thousands of genetic sequences taken from three strains of viruses, influenza, HIV, and Sars-Cov-2. This model was then used to predict the likelihood of sequences generating escape mutations.
4. NLP tools & techniques
The field of NLP encompasses a broad range of techniques and tools that address multiple applications, such as sentiment analysis, machine translation, chatbots, information extraction, etc., to enable computers to understand (NLU), interpret (NLP), and generate (NLG) natural human language.
In drug discovery and development, NLP has a myriad of potential applications that include categorizing entities like genes, and proteins, extracting relationships between these entities to identify potential novel drug targets, predicting drug-drug interactions, increasing the efficiency of patient-trial matching, detecting adverse drug events, streamlining patent tracking and analysis and ensuring regulatory compliance.
Here's a brief overview of some of the key NLP techniques and tools:
4.1. Named Entity Recognition (NER)
Google BERT
BERT (Bidirectional Encoder Representations from Transformers) is an open-sourced neural network-based technique for NLP pre-training that enables anyone to train their own state-of-the-art question answering system. BERT models are able to understand the intent behind Google search queries by seeing a word in the context of the words preceding and following it. The BERT model can be fine-tuned to facilitate state-of-the-art NER. There are also BERT variations, like SpanBERTa for NER.
4.2. Tokenization
NLTK
NLTK (Natural Language Toolkit) is the go-to API for NLP with Python. It enables the pre-processing of text data and helps convert text into numbers for further analysis with ML models. The advantages of word tokenization with NLTK includes white space tokenization, dictionary-based tokenization, rule-based tokenization, regular expression tokenization, Penn treebank tokenization, spacy tokenization, Moses tokenization, and subword tokenization.
TextBlob
TextBlob is a Python (2 and 3) library for processing textual data. A simple API enables users to easily access its methods and perform basic NLP tasks.
spaCy
spaCy is a free, open-source library for advanced NLP in Python. Apart from tokenization, spaCy features a range of capabilities related to linguistic concepts as well as to general machine learning functionality.
Gensim
Gensim is a library for unsupervised topic modelling that also contains a tokenizer.
Keras
Keras is a deep learning API written in Python, running on top of the machine learning platform TensorFlow. Keras’ tokenizer class is used for vectorizing a text corpus.
4.3. Stemming and Lemmatization
Stemming and Lemmatization are NLP algorithms used to normalise text and prepare words and documents for further ML processing. Stemming is used to remove suffixes from similar words to create a word stem that is common to multiple words. This enables NLP models to understand how multiple words are somehow similar. Lemmatization is a progression of Stemming whereby different inflexions of a word are grouped together to be analysed as a unit.
4.4. Bag of Words
Bag of Words is a simple vectorisation technique involving three operations. The input text is first tokenized, then unique words from the tokenized list are selected and alphabetised to create the vocabulary, and finally the frequency of vocabulary words is used to create a sparse matrix.
4.5. Sentence Segmentation
Sentence segmentation, also known as sentence tokenization, is the technique of dividing a string of written language into its component sentences. The key difference between segmentation and tokenization is that the former is a more generic approach to splitting the input text while the latter is performed on the basis of pre-defined criteria.
4.6. Transformers
Since their introduction in 2017, transformers have revolutionized NLP. A multi-head self-attention mechanism, a key component of a transformer architecture, enables the model to focus on different elements of an input sequence, compute attention scores for each element, and then assess their contribution to the final representation. The multi-head attention mechanism enables the model to adapt to different tasks, and self-attention allows for the parallel processing of input sequences, thereby making this approach much faster, more efficient, and more versatile than conventional recurrent or convolutional models. The transformer architecture provided the foundation for the development of multiple models including BERT, LaMDA (Language Model for Dialogue Applications) (LaMDA), and GPT (Generative Pre-Trained Transformer).
4.7. Word Embedding Models
Word embedding is an NLP technique to convert linguistic components, such as words, into numerical representations that machines can comprehend. It is a technique that is central to many NLP applications including text classification, sentiment analysis, NER, etc. Word embedding techniques are broadly categorized into conventional, distributional, and contextual models. Some of the most prominent word embedding models are Term Frequency-Inverse Document Frequency (TF-IDF), Global Vectors for Word Representation (GloVe), and word2vec. Word vectors have also been extended to biological sequence segments, as biovectors, proteins, as Protvec, and gene sequences, as GeneVec.
5. Knowledge graphs
The coinage of the term Knowledge Graphs (KGs) traces back to Austrian linguist Edgar W. Schneider in a 1972 discussion about modular instructional systems. However, as early as the 1950s and 60s, computational linguists were already exploring the possibilities of representing knowledge as relations between concepts in semantic networks. In the 1980s, a cooperative Netherlands university research project, Knowledge Graphs, focused on the design of semantic networks with edges restricted to a limited set of relations to facilitate algebras on the graph. But the modern KG movement really took off with a 2012 Google post, Introducing the Knowledge Graph: things, not strings.
The most recent Gartner Hype Cycle for Artificial Intelligence (AI) identifies knowledge graphs as a must-know innovation within the category of data-centric AI that can be expected to drive high or even transformational benefits.
Definition of knowledge graphs
Despite the ubiquity of knowledge graphs, it still lacks a commonly accepted definition. In 2016, a research project that set out to propose a standard definition of knowledge graphs found that contemporary KG-related research often referenced several contradicting descriptions and misleading assumptions. Following a detailed analysis of KG terminologies and assumptions, the proposed definition was that “A knowledge graph acquires and integrates information into an ontology and applies a reasoner to derive new knowledge”.
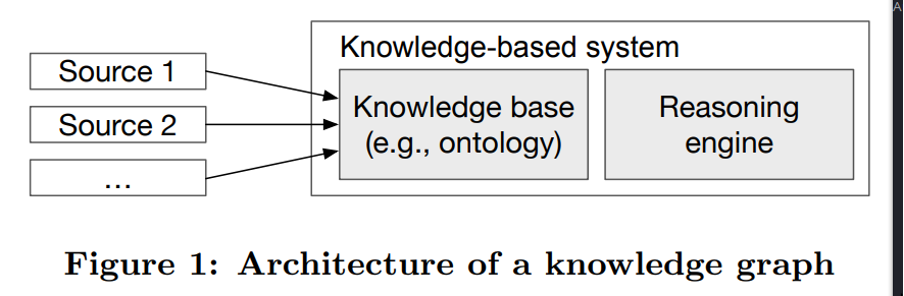
SOURCE: Towards a Definition of Knowledge Graphs
In order to address the issue that knowledge base and knowledge graphs were often used interchangeably, the definition emphasized that KGs must integrate one or more information sources AND have the reasoning capabilities to derive new knowledge. Ergo, a KG with reasoning capabilities but no support for integration was just a knowledge-based system.
Read more on our blog: Knowledge Graphs & The Power Of Context
Key components of a KG framework
As illustrated above, a knowledge graph can be described in terms of two key components - a knowledge database and a reasoning engine. The knowledge database organizes heterogeneous and interconnected data aggregated from different sources into different types of entities, relations, and attributes on the basis of a predefined ontology that evolves along with the KG. The reasoning engine applies various reasoning techniques to automatically infer new knowledge from patterns in the existing data.
However, there are several techniques and technologies that come together in building and updating KGs. Here’s a brief definition of some of the most critical components of most knowledge graph frameworks.
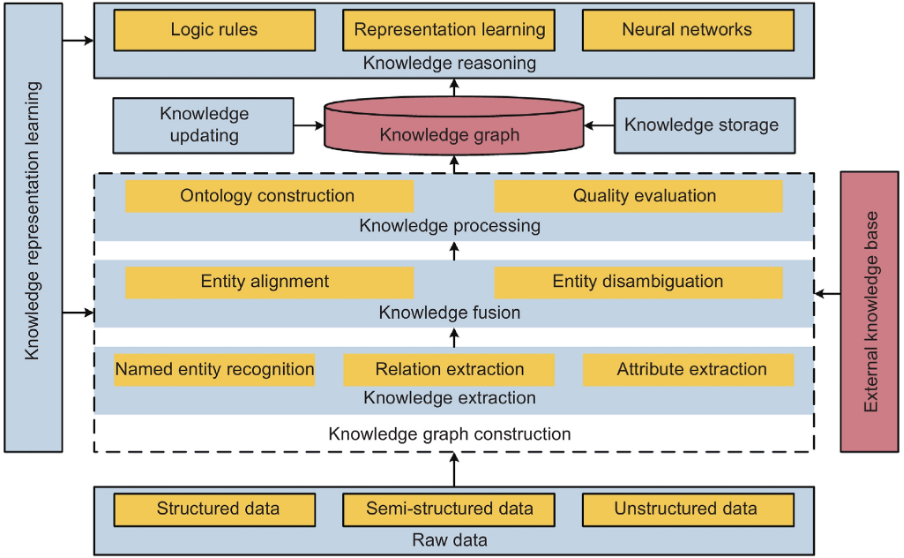
Knowledge Extraction (KE)
Knowledge Extraction is a foundational task in constructing KGs. It refers to the creation of knowledge from structured (relational databases, XML) and unstructured (text, documents, images) sources and representing it in a machine-interpretable format that facilitates inferencing. The main tasks in this phase include named entity recognition (NER), relation extraction (RE), and attribute extraction (AE). There are several methods that are applicable to KE, including rule-based systems, decision trees, and neural networks. However, in the era of large-scale KGs, neural network-based KE is becoming the norm for processing knowledge at scale.
Knowledge Fusion (KF)
Knowledge Fusion is the process of integrating different knowledge graphs into a unified graph. KF ensures semantic interoperability between heterogeneous data sources in order to create a more comprehensive and accurate representation of information. The approach to KF during KG construction is broadly divided into two categories: open source KF, to extract and integrate information from massive fragmented Internet data, and multi-knowledge graph fusion, to merge multiple knowledge graphs based on equivalent instances, classes, and attributes. Key tasks in this process include entity alignment and entity disambiguation.
Knowledge Processing (KP)
Knowledge Processing is the systemization of information in semantic models with common human knowledge in order to create a structured high-quality knowledge graph. Key techniques include ontology construction and quality evaluation.
Knowledge Reasoning (KR)
Knowledge Reasoning is used to predict missing relationships, discover new facts, and update KGs as real-world data evolves. There are two common approaches to KR; one, conventional logic-based deterministic reasoning that yields explainable, interpretable, and transferable results, and two, modern embedding-based reasoning that can deal with uncertainty and data noise, and predict non-determined but plausible knowledge. The current trend is to integrate both approaches for more robust KG reasoning.
Knowledge Representation Learning (KRL)
KRL or KG representation learning (KGRL) is a process that represents entities and relations in a KG as low-dimensional continuous numerical embeddings based on the semantic features of elements. It’s an approach to KG reasoning in which answering a query is reduced to identifying entities with embeddings most similar to the question. Ongoing research efforts to enhance the performance of KRL and therefore the quality of KGs include models for leveraging meta-information for a deeper understanding of semantic features of entities and relations, for integrating multi-modal knowledge, and for querying multi-view KGs.
Knowledge graphs in life sciences
Knowledge graphs may well be the key to tackling many of the AI and data-related challenges in the biomedical and life sciences domain.
Over the past few years, the increasing sophistication of next-generation sequencing (NGS) and multiomics technologies has resulted in the exponential growth of biomedical data. Much of this data is distributed across distributed silos and repositories, a reality that immediately highlights accessibility and interoperability challenges in data integration. Moreover, this distributed data is extremely heterogeneous and includes structured, semi-structured, and multimodal unstructured data, and data schemas/formats can vary based on entity, level of biological organization, domain, technology used, data source, and so on. Unifying multidimensional, multimodal data is the key to characterizing complex biological systems.
Knowledge graphs are increasingly being used to unify vast volumes of data into one systematic model that can elucidate the interrelationships and correlations underlying complex biological systems. Since biomedical data is highly interconnected, it becomes a natural fit for graph representations. for instance, molecular graphs, protein graphs, and compound-level graphs. In addition, the inherent scalability of KGs makes them ideal for at-scale data integration across different structured and unstructured data silos.
Connecting these different layers, types, and sources of biomedical knowledge into one comprehensive knowledge enables biomedical researchers to perform mass-scale cross-comparisons spanning billions of relationships and dependencies to identify unknown patterns and relationships that enable novel insights. Apart from rationalizing data integration and knowledge representation, knowledge graphs will also provide the solid foundation required for AI-driven innovation in biomedical and life sciences research.
Read more on our blog: Biomedical knowledge graphs and the power of ontology
6. NLP challenges in life sciences research
Despite the ever-expanding use cases of NLP in life sciences, there are still several challenges that have to be addressed.
Extracting Biomedical Relationships
Biomedical relation extraction (BioRE), a text mining task to automatically extract and classify relations between different entities in biomedical literature, is critical to many downstream applications, such as literature-based discovery, knowledge graph construction and understanding drug-related interactions. Relation extraction (RE) in biomedical applications is complicated by several challenges including the complexity of biomedical domain-specific vocabulary, the specialized knowledge required to recognize nuances between different entity relationships, and the limited availability of annotated training data. Moreover, even state-of-the-art approaches to BioRE primarily use benchmarking datasets and machine learning models that focus on singular relationships, such as protein-protein interaction. The current focus, therefore, is on automatic annotations and integrating heterogeneous datasets.
Over and above these there are challenges related to linking cross-lingual entities, evaluating semantic relatedness as well as semantic similarity, enhancing the trust and transparency of black-box models, and developing multimodal NLP to integrate heterogeneous data types and thereby address semantic underspecification.
Read more on our blog: Why is NLP challenging?
7. NLP techniques in multi-omics analysis
A core NLP application in multiomics research is in cross-omics data integration. NLP techniques can help extract and normalize entities and relationships across different omics datasets and integrate domain knowledge into that data. However, these techniques are increasingly being used across diverse applications and contexts. Here is a quick overview of some of the most promising:
In biomarker research, NLP has been used variously to identify, extract, and classify biomarker information, in multimodal digital biomarker studies, and in diagnosing cognitive impairment in Alzheimer’s patients using digital voice biomarkers.
In drug discovery and development, NLP technologies have been used to extract drug–drug interactions, detect adverse drug events, and in drug repurposing. In antibody discovery and development, language models have helped perform affinity maturation, create synthetic libraries, and learn antibody features from sequence information.
The list goes much beyond this very short summary of the expanding scope of NLP applications in biomedical research.
8. Future directions and emerging trends in BioNLP
Despite their ability to outperform conventional neural networks in many bioNLP tasks, the view is that new evaluation metrics are needed to fully understand LLM's limitations. A study on biomedical LLMs noted their “encouraging performance” and concluded with the following recommendations:
- The need to develop new LLM-specific data and evaluation paradigms given their poor performance in traditional BioNLP applications, especially named entity recognition.
- A community-wide effort to address the prevalence of errors, missingness, and inconsistencies produced by LLMs prior to deployment.
With that as context, here’s a snapshot of transformative concepts in the field of biomedical NLP:
Generative AI
The fantastic predictions for Generative AI’s impact on the life sciences industry include a 20-40% improvement in productivity, a 2.6-4.5% enhancement in annual pharma revenue, and the potential to raise the efficiency floor and innovation ceiling in drug discovery. In most cases, the optimism comes tempered with the associated risks, concerns, considerations etc.
Knowledge Graphs (KGs)
Knowledge graphs will be foundational to creating the next generation of sophisticated AI systems. KGs will play a key role in capturing data relationships across enterprise-centric heterogeneous models in order to provide AI systems with the context required to make accurate, holistic, and explainable decisions.
Heterogeneous Information Networks (HINs)
HINs are complex networks consisting of multiple types of nodes or edges that can more accurately depict the complicated heterogeneous semantic relations of real-world networks and data. In biomedical research, these multi-layered networks enable the efficient integration of diverse biological data for a more accurate representation of complex biological systems. These networks can not only incorporate diverse entities and relationships but also add important auxiliary information. In recent years, heterogeneous graph neural networks (HGNNs) have emerged to more effectively capture the complex heterogeneity and rich semantics of HIN data.
Vector Databases
Vector databases, aka vector similarity-matching or an approximate nearest neighbor (ANN) service, are designed to store, manage, and search high-dimensional unstructured data. Vector embeddings are numeric representations of data that contain semantic information to help AI systems understand data better. A vector database provides the specialized capabilities required for the storage and querying of vector embeddings.
In Conclusion
- Biomedical NLP continues to expand both in scope and sophistication in parallel with the evolution of NLP technologies & techniques.
- LLMs, the most recent evolutionary milestone in the broader field of NLP, have opened up new opportunities for bioNLP applications. However, risk and accuracy concerns have to be addressed prior to at-scale deployment.
- BioNLP, domain-specific language models derived from general models, still faces the challenges of extensive domain-specific pre-training and fine-tuning before they are ready for biomedical applications.
- Emerging concepts and frameworks, such as vector databases, heterogeneous networks, knowledge graphs, and generative AI, continue to expand the possibilities for NLP applications in biomedical and life sciences research.
9. NLP and BioStrand
NLP technologies are already helping release significant value from vast volumes of unstructured, underutilised textual data in the biomedical domain. However, for a strategic and sustainable impact, NLP technologies have to be organically integrated into conventional omics research processes. A unified framework that can seamlessly combine and analyse all sequence-level, textual and clinical data will be critical to deriving full value from the potential of NLP. And that is the vision driving technology development at BioStrand.
Our unique bottom-up approach to NLP means that researchers now have a solution that is unbiased and domain agnostic. By adding literature analysis to our existing omics and metadata integration framework, we now offer a unified solution that scales across sequence data and unstructured textual data to facilitate a truly integrative and data-driven approach to biological research. Our integrated framework connects omics data/metadata with textual and other unstructured biomedical/clinical data to enable truly integrative research at scale.
Register for future communications:



